MOLECULAR BIOLOGY SKILLS
FROM PROTEIN ENGINEERING TO PURIFICATION
Computational biology, molecular modeling, molecular dynamics, bioengineering
DNA sequencing by Sanger method using radioactive labels and modern automated methods
Protein engineering using bacterial DNA (gene knock-outs, single point mutations, etc)
Protein cloning (PCR, Gibson protocol, etc)
Plasmid vector transformation (electro- and chemical)
Bacterial cell culture growth in various media
Protein expression and co-expression
Protein purification using chromatography (size exclusion, ion exchange, affinity, etc.)
Protein and protein complexes validation (SDS gels, mass-spectrometry, circular dichroism)
Protein complex assembly, proteins renaturation (using buffers and fixatives)
Stoichiometry determination in protein complexes
Immunoblotting

PROTEIN CRYSTALLOGRAPHY
Protein crystallization – manually and using mosquito robot
Crystals visualisation using light microscope and RockImager
Small angle scattering and crystallographic data collection using home rotating anode X-ray source (Rigaku) and at the synchrotrons in France (Soleil, ESRF), Switzerland (SLS) and Germany (DESY)
Data acquisition and processing using HKL 2000, XDS, Mosflm crystallographic software
Structure determination using CCP4, Phenix, Shelx C/D/E, Phaser, ARP/wARP, AutoSharp software

NUCLEAR MAGNETIC RESONANCE
Cloning, expression and purification of C13, N15 isotope labeled proteins
NMR spectroscopy data collection using 600 and 800 MHz Bruker systems
Data acquisition, processing and manual analysis using NMRPipe and Sparky software
Structure determination using NMR software (e.g., AutoProc, AutoAssign) and online servers (DisMeta, Pine)
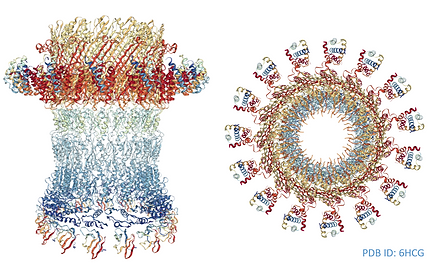
SINGLE PARTICLE CRYO-ELECTRONE MICROSCOPY
Negative stain EM
Protein complex labelling with golden particles
Continuous thin carbon grids preparation
Sample cryo-vitrification using Vitrobot Mark IV (FEI)
Cryo-EM data collection at Titan Krios (Diamond facility, UK) and Tecnai F20 (Imperial College, UK) microscopes
Software usage:
EPU for data collection
MotionCor2 for image frames alignment
GCTF for contrast transfer function dtermination
Relion and IMagic for protein structure reconstruction
LocScale for sharpening of the electron map
I-Tasser, Chimera, Coot, Phenix, Swissmodel for model building and refinement